|
1. Introduction
• Mitochondrial DNA (mtDNA) has become a potent tool in population-, medical-, and forensic genetics investigation. However, the analysis
and comparison of mtDNA sequences may not be easy to do for researchers who are not familiar with mtDNA nomenclature conventions.
• We developed a web program, mtDNAprofiler, which enables users to analyze and compare mtDNA sequences whether they are indicated
as FASTA format or as a difference to revised Cambridge Reference Sequence (rCRS).
• The mtDNAprofiler is composed of four tools; mtDNA nomenclature, mtDNA assembly, mtSNP conversion, and mtSNP comparison.
2. Functions of each tool
| • mtDNA nomenclature tool |

|
|
a. mtDNA nomenclature tool is designed to provide users with automated alignment of mtDNA sequences
and facilitated mtSNP calculation.
b. This tool sequentially performs range determination and mtSNP calculation for the input sequences
followed alignment with the rCRS.
c. Parsimonious Smith-Waterman algorithm is used in this tool as an alignment method.
|
| • mtDNA assembly tool |
|
|
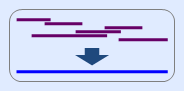
a. mtDNA assembly tool enables to reconstruct the expected sequence from several fragment sequences
obtained from multiple amplification reactions.
b. In particular, this tool is usefull when range of the sequences is included in control region of mtDNA.
c. Even if the sequences are inverted [ex) ATG to CAT], it is no problem to run this tool.
|
| • mtSNP conversion tool |
|
|
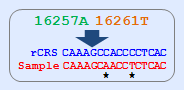
a. mtSNP conversion tool allows mtSNP data indicated as a difference to the rCRS to be converted to
FASTA sequence.
b. This tool basically provides alignment result between rCRS and converted sequences.
c. This tool also allow users to check the validity of mtSNP through the comparison between input and
re-calculated mtSNP data.
|
| • mtSNP concordance-check tool |

|
|
a. mtSNP concordance-check tool is designed to check concordance between two mtSNP data gathered from
independent two experiments for the same sample.
b. In this tool, sequence range of input mtSNP data will be automatically
determined.
c. This tool also allow users to check the validity of mtSNP through the comparison between input and
re-caculated mtSNP data.
|
3. Screen shots
• mtDNA nomenclature tool
• mtDNA assembly tool
• mtSNP conversion tool
• mtSNP concordance check tool
4. Contact us
For comments, bug reports, suggestions for improvement, please contact through e-mail: kjshin@yuhs.ac.
When reporting a bug or any apparent malfunction, please try to include as much information as possible about the problem.
In many cases, the data used help a lot too.
About us, please visit Yonsei DNA Profiling Group.
|